-Search query
-Search result
Showing all 43 items for (author: shi & yf)
EMDB-39012: 
Representative tomogram of primary glioblastoma stem cell with circular inter-mitochondrial junctions.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39015: 
Representative tomogram of microglia cell with nanotunnel-like structures resembling mitochondrial fission.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT
EMDB-39019: 
Representative tomogram of glioblastoma cell with nanotunnel-like structure and inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39021: 
Representative tomogram of normal human astrocyte with nanotunnel-like structure which is an extension of the mitochondrial outer membrane.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39023: 
Representative tomogram of primary glioblastoma differentiated cell with parallel inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39024: 
Representative tomogram of primary glioblastoma stem cell with clustered mitochondria bearing various long-short axis ratios.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-29943: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Yang D, Sheng C, Jinpeng S

EMDB-29944: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Jiangqian M, Sheng C, Jinpeng S

EMDB-29945: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Jianqiang M, Jinpeng S

EMDB-29946: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 3 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Yang D, Shiyi G, Jinpeng S

EMDB-29935: 
Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein
Method: single particle / : Huang SM, Xiong MY, Liu L, Mu J, Sheng C, Sun J

EMDB-29940: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Huang SM, Xiong MY, Liu L, Mu J, Sheng C, Sun J

EMDB-35063: 
The complex structure of Omicron BA.4 RBD with BD604, S309, and S304
Method: single particle / : He QW, Xu ZP, Xie YF

EMDB-29016: 
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Method: single particle / : Zhang J, Shi W, Cai YF, Zhu HS, Peng HQ, Voyer J, Volloch SR, Cao H, Mayer ML, Song KK, Xu C, Lu JM, Chen B
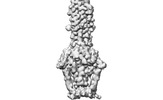
EMDB-29017: 
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Method: single particle / : Zhang J, Shi W, Cai YF, Zhu HS, Peng HQ, Voyer J, Volloch SR, Cao H, Mayer ML, Song KK, Xu C, Lu JM, Chen B

EMDB-29018: 
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Method: single particle / : Zhang J, Shi W, Cai YF, Zhu HS, Peng HQ, Voyer J, Volloch SR, Cao H, Mayer ML, Song KK, Xu C, Lu JM, Chen B

EMDB-34731: 
The structure of MPXV polymerase holoenzyme in replicating state
Method: single particle / : Peng Q, Xie YF, Kuai L, Wang H, Qi JX, Gao F, Shi Y

EMDB-25648: 
CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385
Method: single particle / : Zhang KH, Wu H, Hoppe N, Manglik A, Cheng YF

EMDB-32944: 
The complex structure of Omicron BA.1 RBD with BD604, S309,and S304
Method: single particle / : Huang M, Xie YF
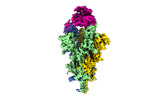
EMDB-32856: 
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein with human ACE2 receptor, C2 state
Method: single particle / : Han WY, Wang YF
EMDB-32857: 
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein with human ACE2 receptor, C3 state
Method: single particle / : Han WY, Wang YF

EMDB-32558: 
Cryo-EM structure of SARS-CoV-2 Omicron spike protein with ACE2, C1 state
Method: single particle / : Han WY, Wang YF

EMDB-32559: 
Cryo-EM structure of Omicron S-ACE2, C2 state
Method: single particle / : Han WY, Wang YF

EMDB-32560: 
Cryo-EM structure of SARS-CoV-2 Omicron spike protein with human ACE2 (focus refinement on RBD-1/ACE2)
Method: single particle / : Han WY, Wang YF

EMDB-32428: 
RBD-1 of SARS-CoV-2 Beta spike in complex with S5D2 Fab
Method: single particle / : Wang YF, Cong Y

EMDB-32430: 
SARS-CoV-2 Beta spike in complex with one S5D2 Fab
Method: single particle / : Wang YF, Cong Y

EMDB-32431: 
SARS-CoV-2 Beta spike in complex with two S5D2 Fabs
Method: single particle / : Wang YF, Cong Y

EMDB-32433: 
SARS-CoV-2 Beta spike in complex with three S5D2 Fabs
Method: single particle / : Wang YF, Cong Y

EMDB-32434: 
SARS-CoV-2 Beta spike SD1 in complex with S3H3 Fab
Method: single particle / : Wang YF, Cong Y

EMDB-32435: 
SARS-CoV-2 Beta spike in complex with three S3H3 Fabs
Method: single particle / : Wang YF, Cong Y

EMDB-32437: 
SARS-CoV-2 Beta spike in complex with two S3H3 Fabs
Method: single particle / : Wang YF, Cong Y

EMDB-32170: 
SARS-CoV-2 Beta variant spike protein in transition state
Method: single particle / : Xu C, Cong Y

EMDB-31542: 
SARS-COV-2 Spike RBDMACSp6 binding to hACE2
Method: single particle / : Wang X, Cao L

EMDB-31543: 
SARS-COV-2 Spike RBDMACSp25 binding to hACE2
Method: single particle / : Wang X, Cao L

EMDB-31544: 
SARS-COV-2 Spike RBDMACSp36 binding to hACE2
Method: single particle / : Wang X, Cao L

EMDB-31546: 
SARS-COV-2 Spike RBDMACSp36 binding to mACE2
Method: single particle / : Wang X, Cao L

EMDB-30702: 
S-2H2-F3a structure, two RBDs are up and one RBD is down, each RBD binds with a 2H2 Fab.
Method: single particle / : Cong Y, Wang YF

EMDB-30703: 
S-2H2-F1 structure, one RBD is up and two RBDs are down, only up RBD binds with a 2H2 Fab
Method: single particle / : Cong Y, Wang YF

EMDB-30704: 
S-2H2-F2 structure, two RBDs are up and one RBD is down, each up RBD binds with a 2H2 Fab.
Method: single particle / : Cong Y, Wang YF

EMDB-30705: 
S-2H2-F3b structure, three RBDs are up and each RBD binds with a 2H2 Fab.
Method: single particle / : Cong Y, Wang YF

EMDB-3125: 
Structural basis for specific recognition of single-stranded RNA by toll-like receptor 13
Method: single particle / : Song W, Wang J, Han ZF, Zhang YF, Zhang HQ, Wang WG, Chang JB, Xia BS, Fan SL, Zhang DK, Wang JW, Wang HW, Chai JJ

EMDB-3026: 
Electron cryo-microscopy of porcine Factor VIII bound to lipid nanotubes and single particle tomography reconstruction
Method: subtomogram averaging / : Dalm D, Galaz-Montoya JG, Miller JL, Grushin K, Villalobos A, Koyfman AY, Schmid MF, Stoilova-McPhie S

EMDB-3027: 
Electron cryo-microscopy of porcine Factor VIII bound to lipid nanotubes and helical reconstruction
Method: helical / : Dalm D, Galaz-Montoya JG, Miller JL, Grushin K, Villalobos A, Koyfman AY, Schmid MF, Stoilova-McPhie S
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
